Welcome!
I am currently a tenure-track
Assistant Professor in the Department
of Mathematics at Kennesaw State University. Prior
to this, I was a Visiting Assistant Profess in the Department
of Mathematics at Virginia Tech. I was formerly an
NSF DIRAC-RTG postdoctoral fellow in the Department of
Applied Mathematics at the University of California,
Merced. In 2022 I was supported by the American Association of
Immunologists (AAI) Intersect Fellowship. My
pronouns are she/her/hers.
I am an applied mathematician working in mathematical
biology. My research lies at the interface of dynamical
systems, numerical analysis, uncertainty quantification,
inverse problems, and their applications to complex
biological systems. In addition to developing and
analyzing mathematical models, I have also gained hands-on
experience in experimental design through collaboration
with biologists.
I co-founded the Math and Computer
Science Research Engagement (MCoRE) group at UC
Merced in Fall 2022, a group aims to foster academic
engagements between UC Merced and the local community in
order to increase diversity and inclusion in STEM fields,
both in industry and academia. MCoRE is currently
supported by a Civic Engagement Microgrant from
Research!America.
About Me
Education
- Ph.D. in Mathematics, University of Louisiana at Lafayette (UL Lafayette), 2012–2017
- B.S. in Fundamental and Applied Mathematics, Beijing University of Posts and Telecommunications (BUPT), 2007–2011
Academic Appointment
- Assistant Professor, Department
of Mathematics, Kennesaw State University,
2024-present
- Visiting Assistant Professor, Department
of Mathematics, Virginia Tech, 2023-2024
- Postdoctoral Fellow, Department of
Applied Mathematics, University of California,
Merced (UC Merced), 2020–2023
– AAI Intersect Postdoctoral Fellow, 2022–2023
– NSF DIRAC-RTG Postdoctoral Researcher, 2020–2022, 2023
– Affiliate Member, Health Sciences Research Institute, 2021–2023 - Postdoctoral Fellow, Institute for Modeling Collaboration and Innovation (IMCI), University of Idaho (UI), 2018–2020
Selected Awards
-
American Institute of Mathematics (AIM) Structured Quartet Research Ensembles (SQuaRE), with Atkins, Case, Huijben, Saucedo, and Ponce; ``Modeling insecticide resistance and management with populations genetics and control theory'', 2023
-
Convergence Accelerator Team Award, with Childs, Edholm, Patterson, Ponce, Prosper, and Qu; the NSF-Simons Center for Multiscale Cell Fate Research, University of California, Irvine (UC Irvine), 2022
-
2022 Intersect Fellowship for Computational Scientists and Immunologists; the American Association of Immunologists (AAI)
-
Career Booster Award, with Omosun; Center for Complex Biological Systems (CCBS), UC Irvine, 2019
-
National First Prize, China Undergraduate Mathematical Contest in Modeling 2010 (CUMCM-2010)
-
National Second Prize, CUMCM-2009
MOOC
- Introduction to Complexity, Santa Fe Institute.
Certificate earned on Dec 4, 2019. [URL]
- Introduction to Dynamical Systems and Chaos, Santa Fe Institute. Certificate earned on Dec 3, 2019. [URL]
- Introduction to Agent-Based Modeling, Santa Fe
Institute. Certificate earned on Sept 17, 2019. [URL]
- R Programming, Johns Hopkins University, Coursera. Certificate earned on June 21, 2019. [URL]
- The Data Scientist’s Toolbox, Johns Hopkins
University, Coursera. Certificate earned on June 10,
2019. [URL]
Media
- "Ordinary
Differential Equation Model Investigates the
Dynamics of Legionnaires' Disease"
- ''Postdoc
Awarded Fellowship to Support Computation and
Immunology Cross-training''
- ''Early-career researchers gear up for virtual Mathematics Research Communities''
Research
I develop and analyze mathematical models
to explain observed experimental dynamics of biological
systems and gain insight into underlying mechanisms. I
believe that mathematical biology at its best requires an
iterative approach in developing mathematical and biological
understanding, in which biological phenomena motivate the
development and refinement of mathematical models, and
mathematical results guide the refinement of empirical
studies. I have used this iterative approach in my research,
and the mathematical techniques I use are tailored to the
specific scenario.
My work has involved both analytical and numerical
approaches, inverse problem methodologies, stochastic and
deterministic modeling frameworks, bioinformatic analysis,
and multi-scale modeling approaches. Below is a list of
selected ongoing projects.
Publications
Peer-Reviewed Journal Articles:
-
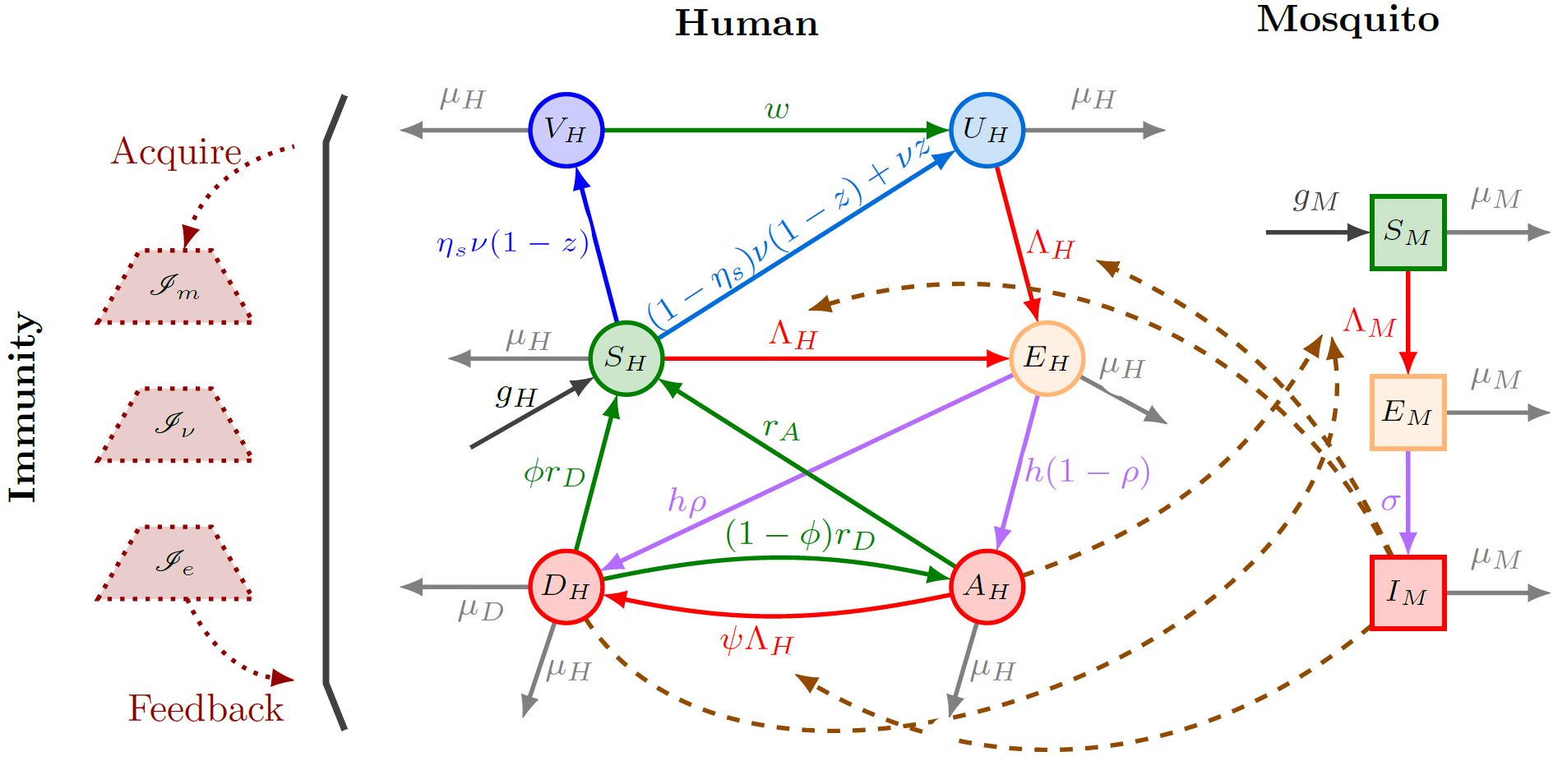
Zhuolin Qu, Denis Patterson, Lihong Zhao, Joan Ponce, Christina J. Edholm, Olivia F. Prosper Feldman, Lauren M. Childs. ``Mathematical modeling of malaria vaccination with seasonality and immune feedback''. PLOS Computational Biology (2025) 21(5): e1012988. doi: 10.1371/journal.pcbi.1012988
-
Mark Wang, Christina J. Edholm, Lihong Zhao. ``Using Mathematical Modeling to Study the Dynamics of Legionnaires’ Disease and Consider Management Options''. Mathematical Biosciences and Engineering (2025) 22(5):1226-1242. doi: 10.3934/mbe.2025045
-
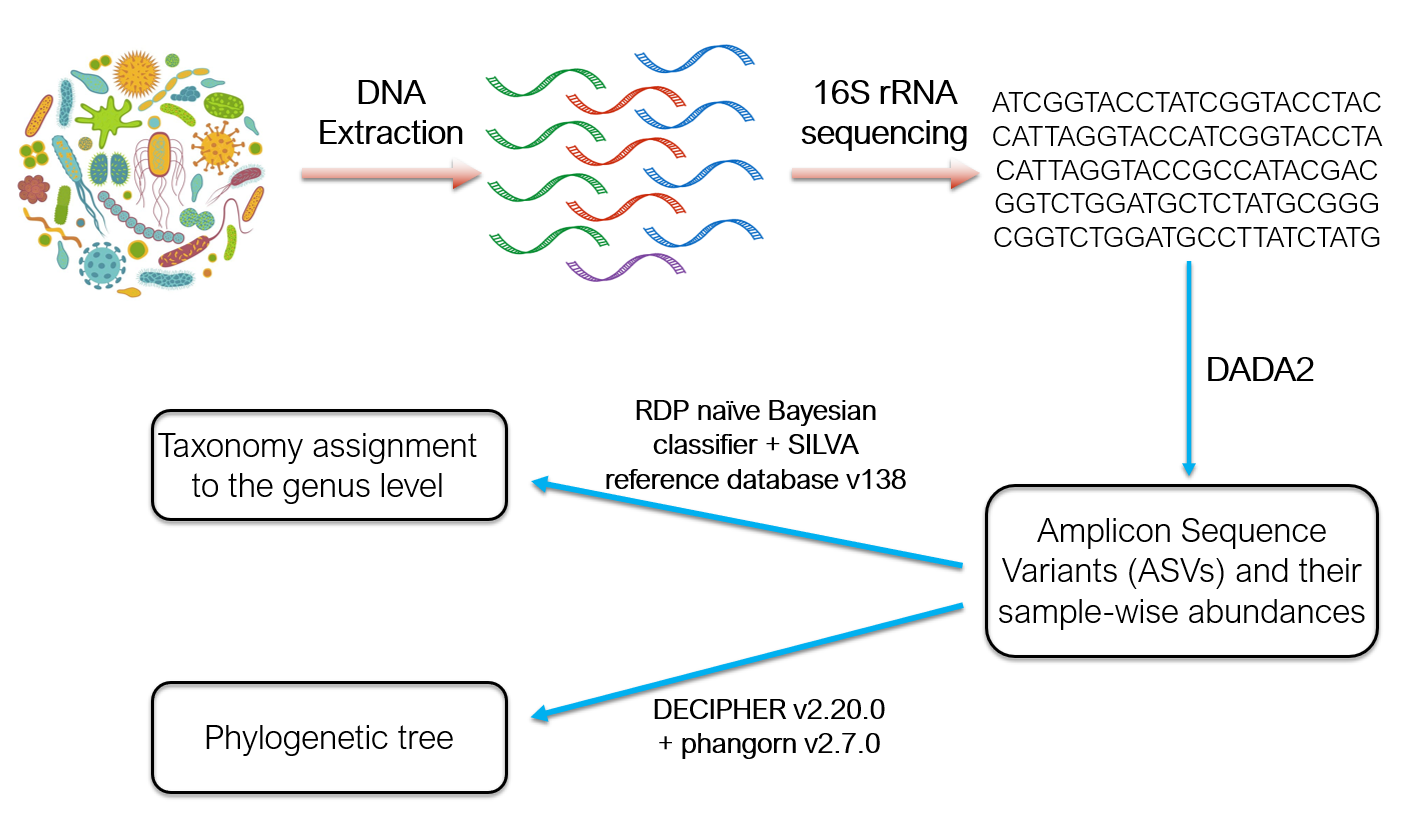
Susana Tejeda-Garibay, Lihong Zhao, Nicholas Hum, Maria Pimentel, Anh L. Diep, Beheshta Amiri, Suzanne Sindi, Dina R. Weilhammer, Gabriela G. Loots, Katrina K. Hoyer. ``Host tracheal and intestinal microbiomes inhibit Coccidioides growth in vitro''. Microbiology Spectrum (2024) e02978-23. doi: 10.1128/spectrum.02978-23
-
Lihong Zhao, Stephanie R. Lundy, Francis O. Eko, Joeseph U. Igietseme, Yusuf O. Omosun. ``Genital tract microbiome dynamics are associated with time of Chlamydia infection''. Scientific Reports (2023) 13:9006. doi: 10.1038/s41598-023-36130-3
-
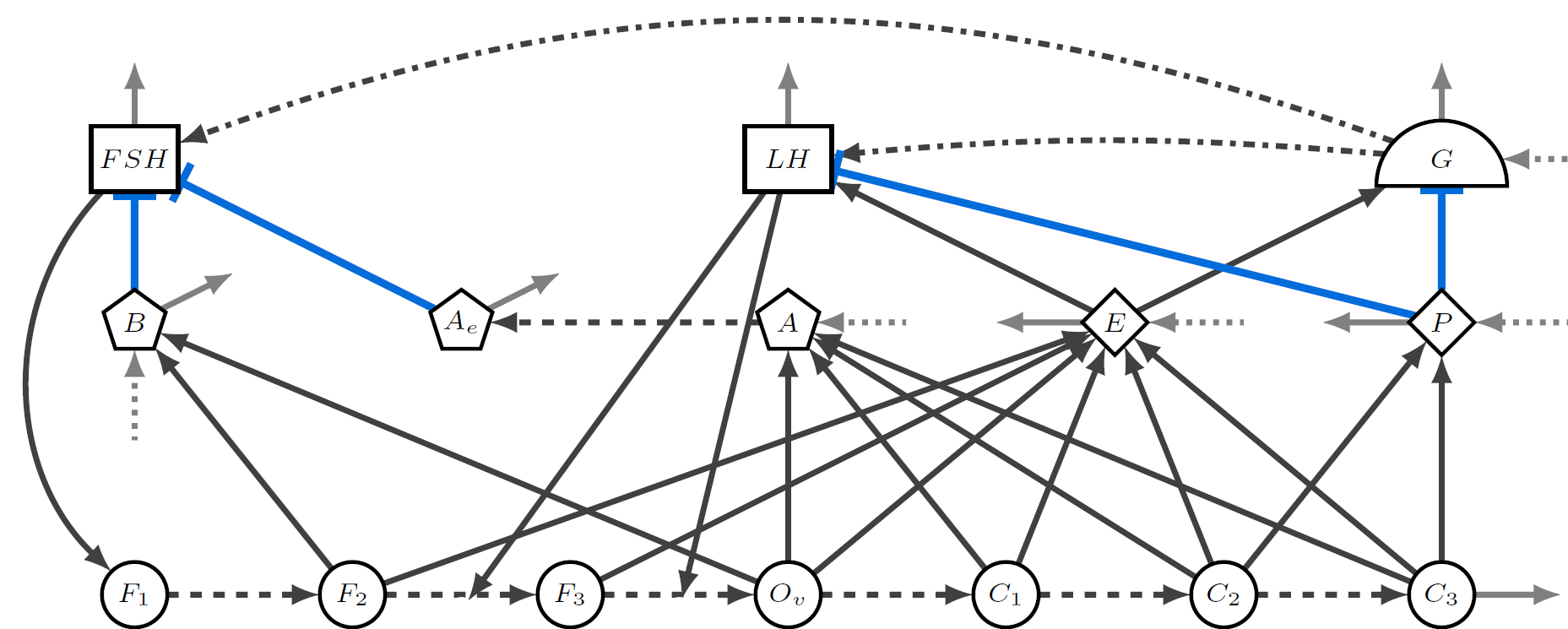
Zhuolin Qu, Denis Patterson, Lauren Childs, Christina Edholm, Joan Ponce, Olivia Prosper, Lihong Zhao. ``Modeling Immunity to Malaria with an Age-Structured PDE Framework''. SIAM Journal on Applied Mathematics (2023) 83(3):1098-1125. doi: 10.1137/21M1464427
-
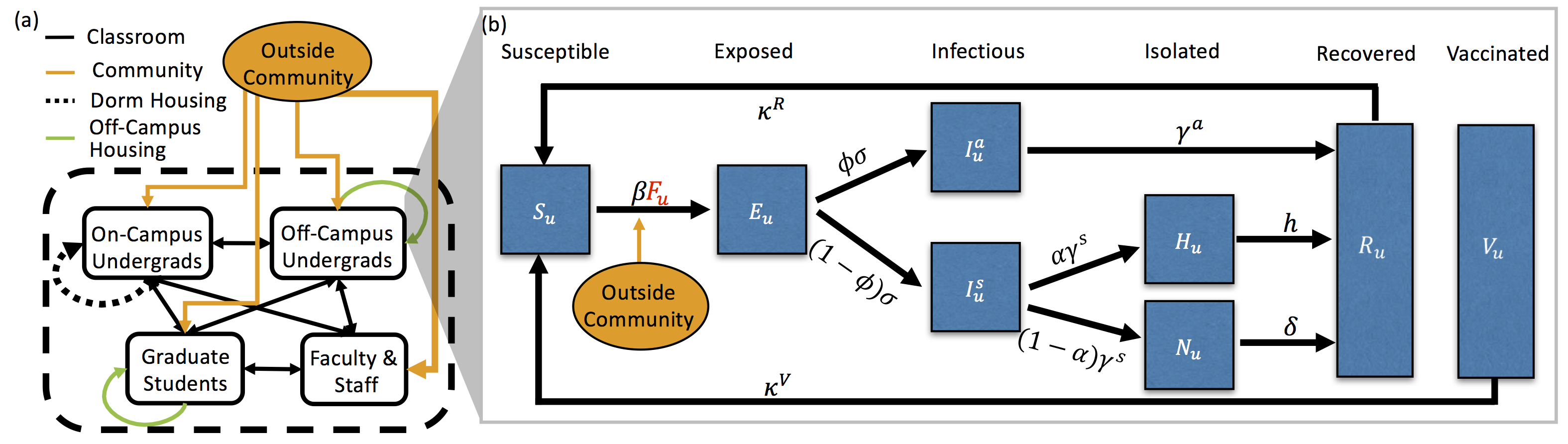
Lihong Zhao, Fabian Santiago, Erica M. Rutter, Shilpa Khatri, Suzanne Sindi. ``Modeling and Global Sensitivity Analysis of Strategies to Mitigate COVID-19 Transmission on a Structured College Campus''. Bulletin of Mathematical Biology (2023) 85:13. doi: 10.1007/s11538-022-01107-2
-
A. Ali Heydari, Oscar A. Davalos, Lihong Zhao, Katrina K. Hoyer, Suzanne S. Sindi. ``ACTIVA: realistic single-cell RNA-seq generation with automatic cell-type identification using introspective variational autoencoders''. Bioinformatics (2022): btac095. doi: 10.1093/bioinformatics/btac095
-
Azmy S. Ackleh, Karyn L. Sutton, Tinging Tang, Lihong Zhao. ``A Second Order Finite Difference Scheme for a Variable Infection-Structured Model of Mycobacterium marinum Dynamics in Aquatic Animals''. Journal of Nonlinear and Variational Analysis (2018) 2(2):177-202. doi: 10.23952/jnva.2.2018.2.06
-
Karyn L. Sutton, Lihong Zhao, Jacoby Carter. ``The Estimation of Growth Dynamics for Pomacea maculata from Hatchling to Adult''. Ecosphere (2017) 8(7): e01840. doi: 10.1002/ecs2.1840
-
Jared T. Guilbeau, Md Istiaq Hossain, Sam D. Karhbet, Ralph Baker Kearfott, Temitope S. Sanusi, Lihong Zhao. ``A Review of Computation of Mathematically Rigorous Lower Bounds on Optima of Linear Integer Programs''. Journal of Global Optimization (2017) 68:677-683. doi: 10.1007/s10898-016-0489-2
Peer-Reviewed Articles in Proceedings:
-
Lihong Zhao, Ruby Kim, Lucy S. Oremland, Mukti Chowkwale, Lisette G. de Pillis, and Heather Z. Brooks. ''A survey of mathematical modeling of hormonal contraception and the menstrual cycle''. Mathematical Modeling for Women’s Health - Collaborative Workshop for Women in Mathematical Biology (eds. Ashlee N. Ford Versypt, Rebecca A. Segal, Suzanne S. Sindi) (2024) Pages 51-82. Cham: Springer Nature Switzerland. doi: 10.1007/978-3-031-58516-6
Other Articles:
-
Lihong Zhao. ``The Vaginal Microbiota and Its Association with Chlamydia''. SIAM News, December 14, 2022. [link]
-
Jared T. Guilbeau, Md Istiaq Hossain, Sam D. Karhbet, Ralph Baker Kearfott, Temitope S. Sanusi, Lihong Zhao. ``Advice for mathematically rigorous bounds on optima of linear programs''. Technical report, University of Louisiana at Lafayette (2016). [pdf]